Flexibilité structurale des biomolécules en milieux complexes
Structural flexibility of biomolecules in complex environmentsHome
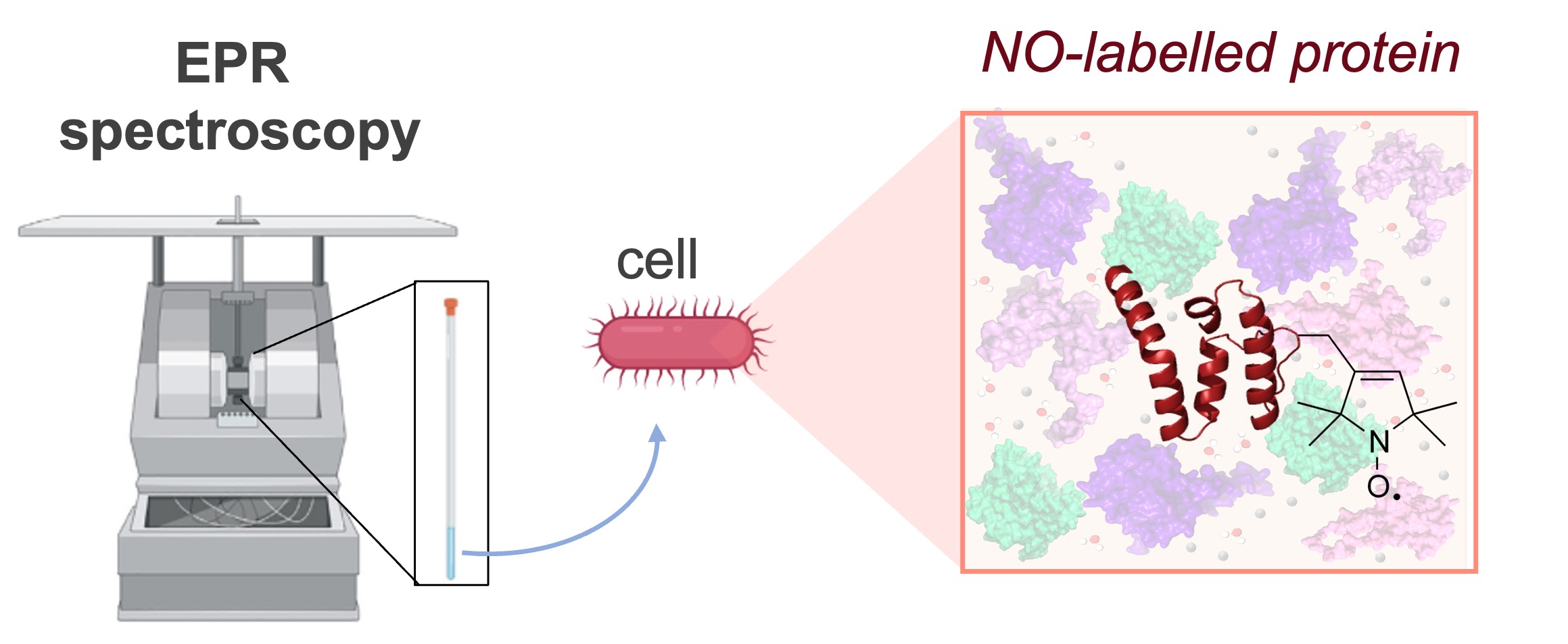
Our team investigates the structural flexibility of biomolecules, with a particular focus on highly flexible proteins, such as molecular chaperones and intrinsically disordered proteins (IDPs). We are also particularly interested in the evaluation of the role of protein dynamics in the biogenesis and the functionalities of large multi-protein complexes. Our research activities are not performed exclusively in simple buffer but also in conditions that closely mimic the cellular environment. Since biophysical studies carry out in vitro often miss key aspects of structural behavior of native environments, our goal is to probe protein function and conformation directly inside living cells.
We develop and apply in-cell EPR spectroscopy combined with Site-Directed Spin Labeling (SDSL) reactions to monitor conformational changes and interactions between biomolecules in real time. A central part of our work consists in the use of nitroxide-based spin labels to modifying the system of interest in order to perform cw- & pulse EPR (i.e. DEER experiments) measurements on a broad panel of flexible proteins. We are also using other types of paramagnetic probes, such as Gd(III)-based spin labels, to carry out orthogonal-SDSL techniques in combination with nitroxide for pulse EPR experiments.
Our team focuses on the development of different aspects of in-cell ERP spectroscopy in order to extend the range of application of these technique in the field of structural biology. We centered our research activities on the use of new classes of nitroxide spin labels suited to perform EPR experiments on proteins in real cellular environments. We are also working to enlarge the range of applicability of this biophysical techniques developing different approaches for the internalization of spin-labelled proteins into cells. At the same time, we are also elaboratin alternative strategies based on the integration of genome-editing techniques and noncanonical amino acids to carry out intracellular labelling reaction and in-cell EPR experiments at physiological conditions.