Biophysique des métalloprotéines et des systèmes dynamiques
Biophysics of metalloproteins and dynamic systemsResearch
Proteic complexes of bacterial energetic metabolism
Prokaryotic molybdenum and tungsten enzymes are involved at key steps of global biogeochemical cycles and contribute at metabolizing various toxic compounds. They participate in biological processes having a strong socioeconomic or environmental interest: denitrification, CO2 production or trapping in relation to the greenhouse effect, cleanup of wasted water and soils, detoxification. In the current research context related to issues of sustainable development, capabilities of molybdenum and tungsten enzymes are an important element for setting up biotechnological processes of great interest.
In this context, we study several bacterial molybdenum and tungsten enzymes, including mainly:
i) nitrate reductases involved in the dissimilatory reduction of nitrate into nitrite, using the periplasmic nitrate reductase (NapAB) from Rhodobacter sphaeroides or the membrane-bound nitrate reductase (NarGHI) from Escherichia coli as model enzymes
ii) formate dehydrogenases catalyzing the reduction of formate into CO2
iii) sulfite oxidases catalyzing the oxidation of sulfite into sulfate, using Thermus thermophilus sulfite dehydrogenase as a model.
We use a multidisciplinary approach combining biochemical and biophysical techniques as well as theoretical modelling to provide an integrated understanding of the catalytic mechanisms of these enzymes, including all constitutive processes: catalysis at the active site, proton and electron transfer, intramolecular transport of substrate or product. We pay a particular attention to identify the molecular factors driving substrate selectivity, reactivity and directionality of the catalysed reaction. This approach is also of great value for understanding evolutionary processes leading to molecular adaptation of these enzymes to changing environments or substrates.
Finally, we work at elucidating at the molecular level the mechanisms that ultimately lead to the formation of active molybdoenzymes during their biosynthesis, with particular attention to the role of dedicated chaperones involved in molybdoenzyme maturation.
Our goal is to provide detailed insights into quinone utilization mechanisms by membrane-bound bioenergetic complexes that couple electron transfer to the generation of a transmembrane proton electrochemical gradient during respiration. In particular, we combine the use of advanced EPR techniques (ENDOR, ESEEM/HYSCORE), redox potentiometry, specific isotopic labeling strategies and computational studies (DFT) to elucidate the molecular factors responsible for tuning the reactivity and specificity of these complexes towards quinones.
Our efforts are mainly directed at the membrane-bound nitrate reductase A (NarGHI) from Escherichia coli, an energy transducing complex optimally induced in anaerobiosis and in high nitrate concentration in the growth medium. Associated to formate dehydrogenase, it forms a paradigmatic redox loop.

NarGHI was known to oxidize ubiquinols (high potential quinones primarily involved in aerobic respiration) and menaquinols (low potential electron carriers primarily involved in prokaryotic anaerobic respiration). In close collaboration with the group of Axel Magalon (Laboratoire de Chimie Bactérienne, CNRS, Marseille), we have shown that NarGHI can also oxidize demethylmenaquinols (a respiratory quinone with intermediate redox potential), and that it stabilizes the paramagnetic semiquinone intermediate of the three respiratory quinones at a single site located within the membrane subunit NarI. Thus, NarGHI constitutes an ideal model system to address at the molecular level the question of the adaptation of an anaerobic enzyme to oxygenic conditions.

Research highlights:
- HYSCORE evidence that mena- and ubisemiquinones bind at the same Q-site (QD) of E. coli nitrate reductase A

Using an E. coli strain deficient in menaquinone biosynthesis, purified NarGHI-enriched inner membrane vesicles were titrated and monitored by EPR spectroscopy, revealing the formation of protein-bound ubisemiquinone species. 2D ESEEM (HYSCORE) experiments on these radicals revealed the same magnetic interaction with a 14N nucleus than that found for the menasemiquinone stabilized at the QD quinol oxidation site of E. coli NarGHI and assigned to His66 Nd, a distal heme axial ligand. This signature is indeed lost in the NarGHIH66Y mutant which is known to be unable to react with quinols. These findings demonstrate that NarGHI-bound USQ can be formed and detected by EPR. They also provide the first direct experimental evidence for similar binding of natural menasemiquinones and ubisemiquinones within the same protein Q-site of NarGHI.
From R. Arias-Cartin, S. Lyubenova, P. Ceccaldi, T. F. Prisner, A. Magalon, B. Guigliarelli & S. Grimaldi*, HYSCORE evidence that mena- and ubisemiquinones bind at the same Q-site (QD) of E. coli nitrate reductase A, J. Am. Chem. Soc. (2010), 132, 5942-5943, https://doi.org/10.1021/ja1009234.
- Probing the menasemiquinone binding mode to nitrate reductase A by selective 2H labelling, HYSCORE spectroscopy and DFT modeling

To directly measure the hyperfine coupling to the methyl protons of MSKD, NarGHI was overproduced in a methionine auxotroph E. coli strain grown in a minimal medium supplemented with 2H-labelled L-methionine at the side chain methyl group. Indeed, the menaquinone methyl group originates from S-adenosylmethionine in the menaquinone biosynthetic pathway. This approach allowed the unambiguous characterization of the 2H – and hence of the 1H – methyl isotropic hyperfine coupling by 2H HYSCORE. The obtained value is the lowest one measured so far for methyl protons of vitamin K molecules (i.e. menadione, menaquinones and phylloquinones) bound to proteins or dissolved in protic solvents. Such results were interpreted by DFT calculations using simple molecular models of the QD site consisting of a menasemiquinone anion hydrogen-bonded to an imidazole ring. They were explained by a strongly asymmetric binding mode of the radical to the protein.
From M. Seif Eddine, F. Biaso, R. Arias-Cartin, E. Pilet, J. Rendon, S. Lyubenova, F. Seduk, B. Guigliarelli, A. Magalon & S. Grimaldi*, Probing the menasemiquinone binding mode to nitrate reductase A by selective 2H & 15N labelling, HYSCORE spectroscopy and DFT modeling, ChemPhysChem (2017), 18, 2704-2714, http://dx.doi.org/10.1002/cphc.201700571.
- 1,2H hyperfine spectroscopy and DFT modeling unveil the demethylmenasemiquinone binding mode to E. coli nitrate reductase A (NarGHI).

The binding mode of the demethylmenasemiquinone (DMSK) intermediate to the EcNarGHI QD quinol oxidation site was analyzed in detail using 1,2H hyperfine (hf) spectroscopy in combination with H2O/D2O exchange experiments and DFT modeling, and compared to the menasemiquinone one bound to the QD site (MSKD) previously studied by us. DMSKD and MSKD are shown to bind in a similar and strongly asymmetric manner through a short (~1.7 Å) H-bond. The origin of the specific hf pattern resolved on the DMSKD field-swept EPR spectrum is unambiguously ascribed to slightly inequivalent contributions from two β-methylene protons of the isoprenoid side chain. DFT calculations show that their large isotropic hf coupling constants (Aiso ~ 12 and 15 MHz) are consistent with both (i) a specific highly asymmetric binding mode of DMSKD and (ii) a near in-plane orientation of its isoprenyl chain at Cb relative to the aromatic ring, which differs by ~ 90° to that predicted for free or NarGHI-bound MSK. Our results provide new insights into how the conformation and the redox properties of different natural quinones are selectively fine-tuned by the protein environment at a single Q site. Such a fine-tuning most likely contributes to render NarGHI as an efficient and flexible respiratory enzyme to be used upon rapid variations of the Q-pool content.
From M. Seif Eddine, F. Biaso, J. Rendon, E. Pilet, B. Guigliarelli, A. Magalon & S. Grimaldi*, 1,2H hyperfine spectroscopy and DFT modeling unveil the demethylmenasemiquinone binding mode to E. coli nitrate reductase A (NarGHI), BBA Bioenergetics (2020), https://doi.org/10.1016/j.bbabio.2020.148203.
Publications :
- Seif Eddine, F. Biaso, J. Rendon, E. Pilet, B. Guigliarelli, A. Magalon & S. Grimaldi* 1,2H hyperfine spectroscopy and DFT modeling unveil the demethylmenasemiquinone binding mode to E. coli nitrate reductase A (NarGHI) Biochim. Biophys. Acta – Bioenergetics (2020)
- M. Seif Eddine, F. Biaso, R. Arias-Cartin, E Pilet, J. Rendon, S. Lyubenova, F. Seduk, B. Guigliarelli, A. Magalon & S. Grimaldi* Probing the menasemiquinone binding mode to nitrate reductase A by selective 2H & 15N labelling, HYSCORE spectroscopy and DFT modeling ChemPhysChem (2017), 18, 2704-2714
- J. Rendon, E. Pilet, Z. Fahs, F. Seduk, L. Sylvi, M. Hajj Chehade, F. Pierrel, B. Guigliarelli, A. Magalon* & S. Grimaldi* Demethylmenaquinol is a substrate of Escherichia coli nitrate reductase A (NarGHI) and forms a stable semiquinone intermediate at the NarGHI quinol oxidation site Biochim. Biophys. Acta – Bioenergetics (2015), 1847, 739-747
- S. Grimaldi, B. Schoepp-Cothenet, P. Ceccaldi, B. Guigliarelli & A. Magalon* The prokaryotic Mo/W-bisPGD enzymes family : a catalytic workhorse in bioenergetics Biochim. Biophys. Acta – Bioenergetics (2013),1827, 1048-1085
- R. Arias-Cartin, S. Grimaldi, P. Arnoux, B. Guigliarelli & A. Magalon* Cardiolipin binding in bacterial respiratory complexes : Structural and functional implications Biochim. Biophys. Acta – Bioenergetics (2012), 1817, 1937-1949
- S. Grimaldi*, R. Arias-Cartin, P. Lanciano, S. Lyubenova, R. Szenes, B. Endeward, T. F. Prisner, B. Guigliarelli & A. Magalon Determination of the proton environment of high stability menasemiquinone intermediate in Escherichia coli nitrate reductase A by pulsed EPR J. Biol. Chem. (2012), 287, 4662-4670
- R. Arias-Cartin, S. Grimaldi, J. Pommier, P. Lanciano, C. Schaefer, P. Arnoux, G. Giordano, B. Guigliarelli & A. Magalon* Cardiolipin-based respiratory complex activation in bacteria Proc. Nat. Acad. Sci. USA (2011), 108, 7781-7786
- F. MacMillan*, S. Kacprzak, P. Hellwig, S. Grimaldi, H. Michel, & M. Kaupp Elucidating mechanisms in haem copper oxidases : The high-affinity QH binding site in quinol oxidase as studied by DONUT-HYSCORE spectroscopy and density functional theory Faraday Discussions (2011), 148, 315-344
- R. Arias-Cartin, S. Lyubenova, P. Ceccaldi, T. F. Prisner, A. Magalon, B. Guigliarelli & S. Grimaldi* HYSCORE evidence that mena- and ubisemiquinones bind at the same Q-site (QD) of E. coli nitrate reductase A J. Am. Chem. Soc. (2010), 132, 5942-5943
- S. Grimaldi*, R. Arias-Cartin, P. Lanciano, S. Lyubenova, B. Endeward, T. F. Prisner, A. Magalon & B. Guigliarelli Direct evidence for nitrogen ligation to the high-stability semiquinone intermediate in E. coli nitrate reductase A J. Biol. Chem. (2010), 285, 179-187
- P. Lanciano, A. Magalon, P. Bertrand, B. Guigliarelli & S. Grimaldi* High-stability semiquinone intermediate in the nitrate reductase A (NarGHI) from Escherichia coli is located in a quinol oxidation site close to heme bD Biochemistry (2007), 46, 5323-5329
- S. Grimaldi*, P. Lanciano, P. Bertrand, F. Blasco, & B. Guigliarelli Evidence for an EPR-detectable semiquinone intermediate in the membrane-bound subunit NarI of nitrate reductase A (NarGHI) from Escherichia coli Biochemistry (2005), 44, 1300-1308
Iron sulfur cofactors
Heme and Copper containing enzymes
Oxidative heme enzymes and related natural/artificial proteins
A NOVEL PERIPLASMIC PROTEIN INVOLVED IN COPPER RESISTANCE

The team of Dr. Soufian Ouchane (Department of Microbiology, I2BC, Gif-sur-Yvette) has discovered a new protein involved in copper resistance in the purple photosynthetic bacterium Rubrivivax gelatinosus. This bacterium can grow in a medium containing up to 1.2 mM in copper ions. Under these conditions, it naturally overexpresses a protein, CopI, located in the periplasm. In collaboration with this team, we endeavor to gain insights into this enzyme by using biophysical methods. EPR results show that there are two Cu(II) sites per protein, one of which being a green type cupredoxin. We are currently characterizing both sites to determine their ligands, structural and electronic properties. Since the function of this new protein is unknown, we are also trying to determine possible catalysis and we are studying electron transfer within this system.

EPR spectrum of a frozen solution of purified CopI protein complemented in Cu(II) (bottom). The two top spectra show the deconvolution into both components, a green cupredoxin type signal (top) and a square planar site (middle) that we suppose located in the N terminal His rich portion of the protein. Grey lines represent simulated spectra.
Protein Dynamics
Understanding protein structural dynamics is a critical issue to decipher protein function, their biological activity and mode of interaction with their partners.
Among magnetic resonances, Site-Directed Spin Labeling (SDSL) coupled to Electron Paramagnetic Resonance (EPR) spectroscopy is part of the toolbox available to investigate protein structure and dynamics. It is an accurate and powerful approach to study structural and conformational changes in soluble but also membrane proteins and has demonstrated advantageous features to capture protein dynamics inside cells.
In our lab, we are actively involved in the investigation of flexible and globular protein by SDSL-EPR as well as in the development of the SDSL-EPR approach by working on the design of new spin-labels, non-canonical amino acids and in-cell EPR.
Site-Directed Spin Labeling (SDSL) combined to Electron Paramagnetic Resonance (EPR) is a powerful technique used since the 90s to study structural modification of proteins in solution. The technique is based on the introduction of a spin label (usually a nitroxide) on a specific position (usually introduced by mutagenesis) in the protein.
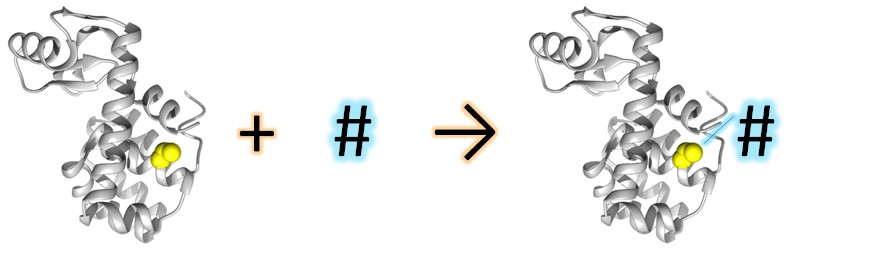
The power of the technique resides on the relation of the EPR spectral shape of the label and its mobility. The EPR spectral shape is very sensitive to the label local environment, this later being related to its structuration. Following the spectral shape evolution allows us to get information on the structural environment of the label and its modification.

Distance measurements
When two paramagnetic sites (i.e. exogenous or endogenous like metal centers) are present on a protein, or in one protein and its partner, one can measure inter-label distances. This is possible using pulsed EPR and DEER (Double Electron Electron Resonance) sequence. The classical range of distances one can reach in biological systems is from ~2 nm up to 8 nm. Distance measurements can then be obtained between two nitroxide labels, one nitroxide label and one natural cofactor like a metal center (Cu2+, Gd3+, Mn2+ …), radical cofactor, aminoacid radical, ….
We used this advanced EPR technique to study the 1-Aminocyclopropane-1-carboxylic Oxidase (ACCO), a non-heme Fe(II) enzyme. Combined with MD studies, we showed that the two conformations (so-called “open” and “closed” conformations) obtained by 1) XR crystallography or 2) in-silico model do not represent what is going on in solution. This picture is probably too simple to explain the enzymatic-function complexity, showing once again that proteins are dynamic with inherent flexibility allowing them to interact with different molecules as substrates, co-factors or partners.

Publications:
Marlène Martinho, Eugénie Fournier, Nolwenn Le Breton, Elisabetta Mileo, Valérie Belle.
Nitroxide spin labels: fabulous spy spins for biostructural EPR applications.
Electron Paramagnetic Resonance, 2019, 26, 66 – 88.
DOI: 10.1039/9781788013888-00066
HAL: hal-01953977v1
Eugénie Fournier, Sybille Tachon, Nicholas J. Fowler, Guillaume Gerbaud, Pascal Mansuelle, Pierre Dorlet, Sam P. De Visser, Valérie Belle, A. Jalila Simaan, Marlène Martinho.
The Hunt for the Closed Conformation of the Fruit‐Ripening Enzyme 1‐Aminocyclopropane‐1‐carboxylic Oxidase: A Combined Electron Paramagnetic Resonance and Molecular Dynamics Study.
Chem. Eur. J., 2019, 25 (60), 13766-13776.
DOI: 10.1002/chem.201903003
HAL: hal-02303326v1
Eugénie Fournier, Elisabetta Mileo, Emilien Etienne, Guillaume Gerbaud, Valérie Belle, Marlène Martinho.
Spin labels: spies in the heart of proteins.
Actualité Chimique, 2019, 443. HAL: (chercher)
Intrinsically Disordered Proteins (IDPs)
For more than 15 years, we have used SDSL technique and EPR to study proteins lacking secondary structure in solution, like Intrinsically Disordered Proteins (IDPs). This approach allowed us to highlight induced folding in IDPs and biological behaviors like protein-protein interaction.
Intrinsically Disordered Proteins (IDPs) constitute a large family of proteins lacking a well-defined 3D structure under physiological conditions, while being involved in many key biological processes. For more than 15 years, we develop and apply Site-Directed Spin Labeling combined with EPR spectroscopy (SDSL-EPR) to characterize such highly flexible biological systems.
An example of IDP is the Tau protein, a microtubule (MTs)-associated protein. Despite extensive studies, the fine structural characterization of Tau in interaction with MTs still remains challenging. Our studies using SDSL-EPR revealed the existence of a thiol-disulfide exchange reaction between Tau and MTs. This system constitutes a typical example of a fuzzy complex, a characteristic that has been proposed to be functionally important by contributing to finely tune binding affinities in protein-protein interaction.
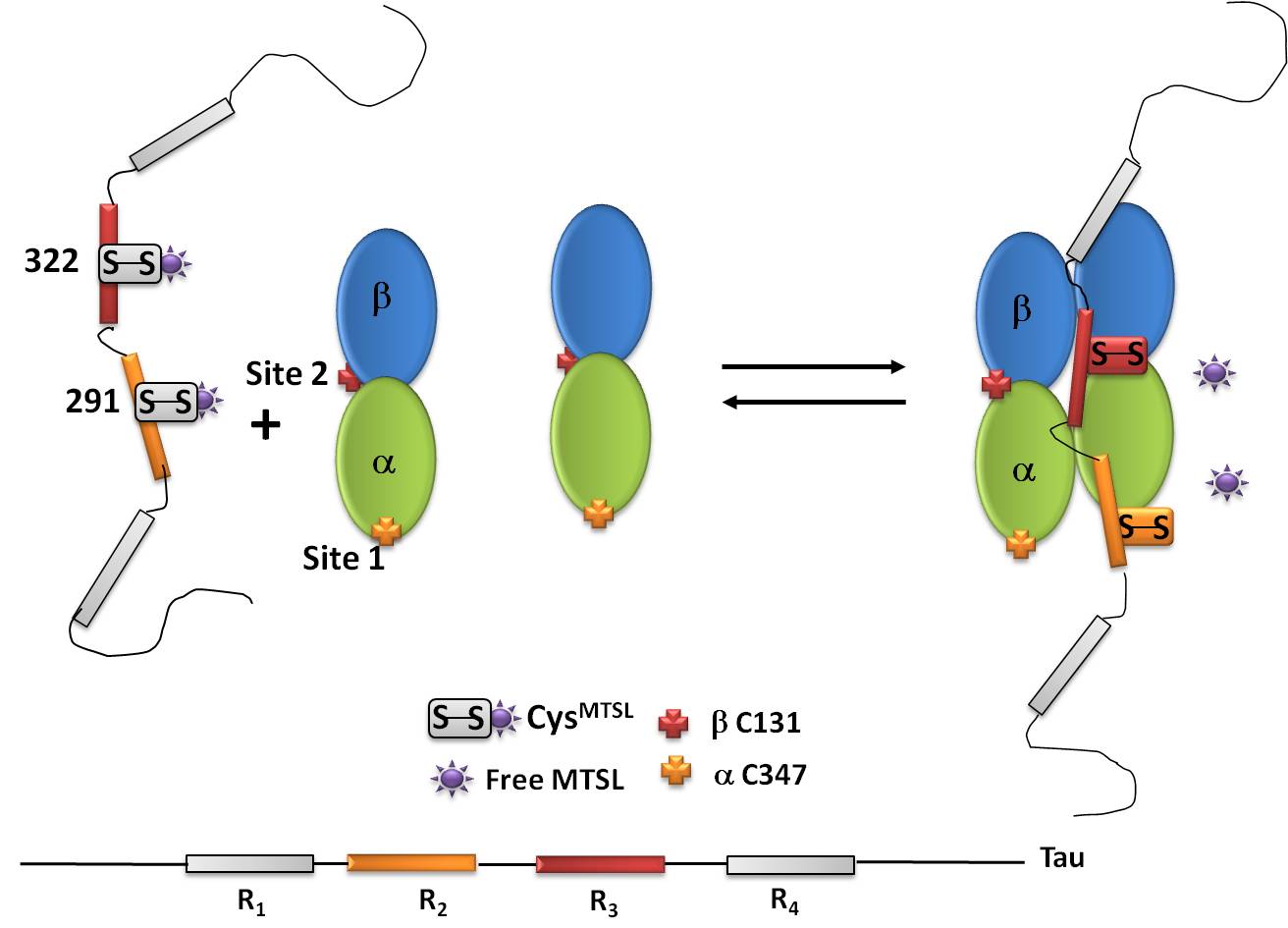
Protein-protein interactions
SDSL-EPR technique allowed us to highlight biological behaviors like protein-protein interaction. We applied this technique on the study of the nucleoprotein (N) of Measles Virus and of Henipa Viruses within the Paramyxoviridae family. The unstructured Cterm region of the N protein, called Ntail, interacts with its partner, namely XD (X domain of the phosphoprotein partner), through an a-helical induced folding. The precise cartography of this interaction has been shown by SDSL for different viruses (Measles and Henipa viruses). Our studies allowed us to specify the structural models of NTAIL–PXD complexes in the viruses proposed previously.

Membrane proteins
SDSL-EPR spectroscopy is a powerful technique that can be applied to membrane proteins without size or environment limitations. It gives information about dynamics, water accessibility and site-specific environment of nitroxide probes.
ATP-Binding Cassette (ABC) transporters use the energy of ATP binding and hydrolysis to function. ABC transporters are a large and ubiquitous superfamily of proteins involved in the translocation of a wide variety of substrates across biological membranes, including amino acids, peptides, sugars, lipids, porphyrins or multiple drugs. Despite a rapid increase in the number of structures of ABC transporters available, their functioning mechanism is still unclear, the only consensus being that transport occurs via an alternating access mechanism. Detailed molecular mechanisms of drug recognition and translocation remain unclear and differ among transporters, the catalytic step (ATP-binding or ATP hydrolysis) responsible for drug translocation is still debated, and the molecular model for transport (switch or constant contact models) is still controversial. In our group, we use SDSL-EPR spectroscopy to study the Bacillus subtilis multidrug transporter BmrA to decipher how it is capable to efflux multiple drugs.
Publications:
Marlène Martinho, Diane Allegro, Isabelle Huvent, Charlotte Chabaud, Emilien Etienne, Hervé Kovacic, Bruno Guigliarelli, Vincent Peyrot, Isabelle Landrieu, Valérie Belle, Pascale Barbier.
Two Tau binding sites on tubulin revealed by thiol-disulfide exchanges.
Sci Rep, 2018, 8:13846, 1 – 11.
DOI: 10.1038/s41598-018-32096-9
HAL: hal-01883844v1
Nolwenn Le Breton, Marlène Martinho, Elisabetta Mileo, Emilien Etienne, Guillaume Gerbaud, Bruno Guigliarelli, Valérie Belle.
Exploring intrinsically disordered proteins using site-directed spin labeling electron paramagnetic resonance spectroscopy.
Front. Mol. Biosci., 2015, 2:21, 1 – 7.
DOI: 10.3389/fmolb.2015.00021
HAL: hal-01429982v1
Elisabetta Mileo, Magali Lorenzi, Jenny Erales, Sabrina Lignon, Carine Puppo, Nolwenn Le Breton, Emilien Etienne, Sylvain R. A. Marque, Bruno Guigliarelli, Brigitte Gontero, Valérie Belle.
Dynamics of the intrinsically disordered protein CP12 in its association with GAPDH in the green alga Chlamydomonas reinhardtii: a fuzzy complex.
Mol. BioSyst., 2013, 1 (11), 349.
DOI: 10.1039/c3mb70190e
HAL: hal-01460463v1
Jenny Erales, Magali Lorenzi, Régine Lebrun, André Fournel, Emilien Etienne, Carine Courcelle, Bruno Guigliarelli, Brigitte Gontero, Valérie Belle.
A New Function of GAPDH from Chlamydomonas reinhardtii: A Thiol−Disulfide Exchange Reaction with CP12.
Biochemistry, 2009, 48 (25), 6034 – 6040.
DOI: 10.1021/bi900569h
HAL: hal-00429826
Marlène Martinho, Johnny Habchi, Zeina El Habre, Léo Nesme, Bruno Guigliarelli, Valérie Belle, Sonia Longhi.
Assessing induced folding within the intrinsically disordered C-terminal domain of the Henipavirus nucleoproteins by site-directed spin labeling EPR spectroscopy.
J. Biomol. Struct. Dyn., 2012, 31, 453-471.
DOI: 10.1080/07391102.2012.706068
Valérie Belle, Sabrina Rouger, Stéphanie Costanzo, Elodie Liquière, Janez Strancar, Bruno Guigliarelli, André Fournel, Sonia Longhi.
Mapping α-helical induced folding within the intrinsically disordered C-terminal domain of the measles virus nucleoprotein by site-directed spin-labeling EPR spectroscopy.
Proteins, 2008, 73 (4), 973 – 988.
DOI: 10.1002/prot.22125
New labels
Commercial nitroxide labels (MTSL, MA-Proxyl, IA-Proxyl, …) are usually designed to be specific of cysteine residues. In order to target other amino acid residues, we developed other nitroxyde labels specific of tyrosine residues:

To go further on the study of protein-protein interactions, we developed a nitroxide label, i.e. phosphorylated Proxyl, having a different spectral signature to be able to follow structural modifications of a protein and its partner at the same time.

Publications
Magali Lorenzi, Carine Puppo, Régine Lebrun, Sabrina Lignon, Valérie Roubaud, Marlène Martinho, Elisabetta Mileo, Paul Tordo, Sylvain R. A. Marque, Brigitte Gontero, Bruno Guigliarelli, Valérie Belle.
Tyrosine-Targeted Spin Labeling and EPR Spectroscopy: An Alternative Strategy for Studying Structural Transitions in Proteins.
Angew. Chem. Int. Ed., 2011, 50, 9108 – 9111.
DOI: 10.1002/anie.201102539
HAL: hal-00677247v1
Elisabetta Mileo, Emilien Etienne, Marlène Martinho, Régine Lebrun, Valérie Roubaud, Paul Tordo, Brigitte Gontero, Bruno Guigliarelli, Sylvain R. A. Marque, Valérie Belle.
Enlarging the Panoply of Site-Directed Spin Labeling Electron Paramagnetic Resonance (SDSL-EPR): Sensitive and Selective Spin-Labeling of Tyrosine Using an Isoindoline-Based Nitroxide.
Bioconjugate Chem., 2013, 24 (6), 1110 – 1117.
DOI: 10.1021/bc4000542
HAL: hal-01460460v1
Nolwenn Le Breton, Marlène Martinho, Kuanysh Kabytaev, Jérémie Topin, Elisabetta Mileo, David Blocquel, Johnny Habchi, Sonia Longhi, Antal Rockenbauer, Jérôme Golebiowski, Bruno Guigliarelli, Sylvain R. A. Marque, Valérie Belle.
Diversification of EPR signatures in site directed spin labeling using a β-phosphorylated nitroxide.
Phys. Chem. Chem. Phys., 2014, 16, 4202 – 4209.
DOI: 10.1039/c3cp54816c
HAL: hal-01460516v1
Christoph Gmeiner, Daniel Klose, Elisabetta Mileo, Valérie Belle, Sylvain R. A. Marque, Georg Dorn, Frédéric H. T. Allain, Bruno Guigliarelli, Gunnar Jeschke, Maxim Yulikov.
Orthogonal Tyrosine and Cysteine Site-Directed Spin Labeling for Dipolar Pulse EPR Spectroscopy on Proteins.
J. Phys. Chem. Lett., 2017, 8 (19), 4852 – 4857.
DOI: 10.1021/acs.jpclett.7b02220
HAL: hal-01596569v1
Nolwenn Le Breton, Sonia Longhi, Antal Rockenbauer, Bruno Guigliarelli, Sylvain R. A. Marque, Valérie Belle, Marlène Martinho.
Probing the dynamic properties of two sites simultaneously in a protein–protein interaction process: a SDSL-EPR study.
Phys. Chem. Chem. Phys., 2019, 21, 22584 – 22588.
DOI: 10.1039/c9cp04660g
HAL: hal-02341622v1
Non-canonical aminoacids
Conventionally, the paramagnetic probes (nitroxides) are specific for cysteines. However, for some systems, chemical modification of these residues is deleterious since they can be important in catalysis and greatly perturb both the function and/or the structure. The incorporation of non canonical amino acids (ncAAs) or non natural amino acids (nnAAs) and their labeling with specific nitroxides has been shown to be a powerful alternative. It is also possible to incorporate two nnAAs on the same protein in order to measure distances between paramagnetic centers. This approach has been expanding in recent years. It uses a pair of non-native suppressor tRNAs and aminoacyl tRNA synthetase to incorporate the unnatural amino acid into the recombinant protein in response to a unique STOP AMBER codon (TAG) . Of all non-amino acids natural which have been genetically coded, some contain a reactive chemical group which can be used to introduce biophysical probes (fluorescence, NMR or EPR), in particular by reactions of bioorthogonal conjugation, click chemistry or by photo activation.

In the lab, we develop these approaches on NADPH cytochrome P450 reductase (CPR) a multidomain protein bearing two individual redox domains: a FMN domain and a FAD domain. Large conformational changes exist in CPR, enabling the FMN domain to move out of the FAD domain cavity and be accessible to various electrons acceptors. CPR exists in two physical states in rapid exchange: a locked state, in which the domains present a compact, closed conformation, and the unlocked state, in which the two domains do not present any interface and move almost independently. Flavins reduction and the presence of cofactors trigger the opening and closing mechanisms, but the conformational space explored in these states is extremely restricted and may not represent the complete picture of the necessary movements when acceptors are present. Therefore we still do not fully understand how intrinsic (CPR molecular determinants, redox state) and extrinsic (ligands, acceptors) factors contribute to the control of the conformational equilibrium. We also hypothesize large conformational changes are triggered/controlled by electron transfer (ET) steps to acceptors.

In-cell EPR
In-Cell EPR allows us to obtain EPR data from proteins in living cells. This kind of studies involves taking in account the effects of the intracellular environment on protein stability, structure, dynamics and protein-protein interactions.
The intracellular environment is extremely heterogeneous and complex. The viscosity of the cytoplasm, the macromolecular crowding, specific and non-specific weak interactions, as well as a myriad of potential interactors constitute parameters that can have a huge impact on protein structure, dynamics, and protein–partner interactions.
Therefore, obtaining a deep understanding of protein function requires to probe structural dynamics of proteins and protein interactions in their native environment, that is, inside cells.

To this aim, in the lab, we have settled up the “in-cell EPR” approach: the Site-Directed Spin Labeling – EPR (SDSL-EPR) and the design of new spin labels (see M-TETPO) are synergically employed to explore proteins in the context of their intracellular environment and in physiological conditions (solution-state and temperature).
The EPR investigation “in-cell” is supported by other biophysical methods (fluorescence microscopy, CD) and by cellular and biochemical studies.
We are currently investigating protein dynamics in bacteria, X. laevis oocytes and eukaryotic cells.
Publications
Ganesan Karthikeyan, Alessio Bonucci, Gilles Casano, Guillaume Gerbaud, Sébastien Abel, Virginie Thomé, Laurent Kodjabachian, Axel Magalon, Bruno Guigliarelli, Valérie Belle, Olivier Ouari, Elisabetta Mileo.
A Bioresistant Nitroxide Spin Label for In-Cell EPR Spectroscopy: In Vitro and In Oocytes Protein Structural Dynamics Studies.
Angew. Chem. Int. Ed., 2018, 57, 1366 – 1370.
DOI: 10.1002/anie.201710184
HAL: hal-02000559v1
Alessio Bonucci, Olivier Ouari, Bruno Guigliarelli, Valérie Belle, Elisabetta Mileo.
In‐Cell EPR: Progress towards Structural Studies Inside Cells.
ChemBioChem, 2020, 21 (4), 451 – 460.
DOI: 10.1002/cbic.201900291
HAL: hal-02180300v1
