Evolution de la bioénergétique
Evolution of bioenergeticsResearch
Energetics
We study selected prokaryotic energy conserving chains with the aim of contributing to a comprehensive picture of the diversity of these chains but also of elaborating common features and conserved principles.
The ultimate goal of this approach is to contribute to the elucidation of the origin(s) and evolutionary pathways of biological energy conversion.
All processes in living cells are energy-dependent. Apart from the comparatively inefficient fermentation pathway, this energy is invariably provided by the « Mitchellian » chemiosmotic mechanism, that is, by the conversion of a proton-motive-force (pmf) across a « bioenergetic membrane » into ATP-synthesis via the enzyme ATPase.
In the vast majority of cases, the pmf is produced by the coupling of electron transport to the vectorial translocation of positively charged ions (generally protons, H+) across the bioenergetic membrane (Figure 1). In a few archaeal species, the bioenergetic membrane potential is alternatively built up directly by light-driven ion-translocation performed by the enzyme Bacteriorhodopsin.

Life thus mainly draws its energy from collapsing electrochemical disequilibria of redox-active substrates. Such disequilibria may be provided by geo-/biochemical processes in the environment or induced by life itself via photochemical mechanisms (i.e. chlorophyll-based photosynthesis).
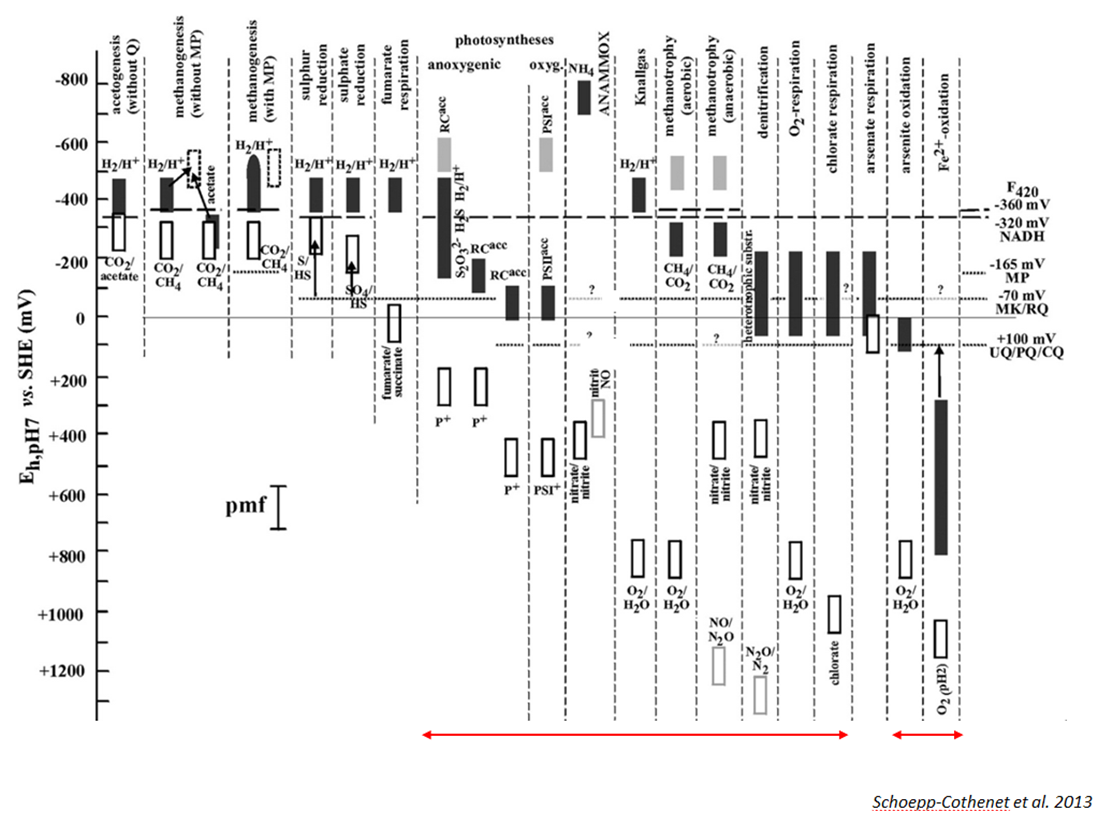
In eukaryotes, the corresponding electron transfer chains are almost invariably aerobic respiration (in mitochondria) or oxygenic photosynthesis (in chloroplasts). Prokaryotes, by contrast, feature a bewildering diversity of bioenergetic electron transfer chains and are probably able to use all bio-available redox compounds as substrates for energy conversion, either as reductants or oxidants. Figure 2 represents a few selected examples of this -probably far from exhaustively known- diversity of prokaryotic electron transfer chains.
BIP09 studies selected prokaryotic energy conserving chains with the aim of contributing to a comprehensive picture of the diversity of these chains but also of elaborating common features and conserved principles. The ultimate goal of this approach is to contribute to the elucidation of the origin(s) and evolutionary pathways of biological energy conversion
Electron Bifurcation (EB)
The molecular process of electron bifurcation (EB) couples the endergonic 1-electron-reduction of a low-potential acceptor by two-electron redox compounds such as flavins or quinones to the exergonic 1-electron-transfer involving the other electron towards a relatively high-potential acceptor molecule. This mechanism represents one of the fundamental principles of free energy conversion in bioenergetics whose basic functional principles still aren’t fully understood (10.3389/fmicb.2018.01357).
Our group has been working on quinone-based electron bifurcation (QBEB) in bc1 and b6f complexes for more than 20 years and more recently also started to investigate this mechanism performed by flavin-containing enzymes, which is an emergent research subject with the first examples reported only a little over 10 years ago (10.1128/JB.01422-07; 10.1128/JB.01417-07).
The electron-bifurcating hydrogenase (HydABC) from Thermotoga maritima
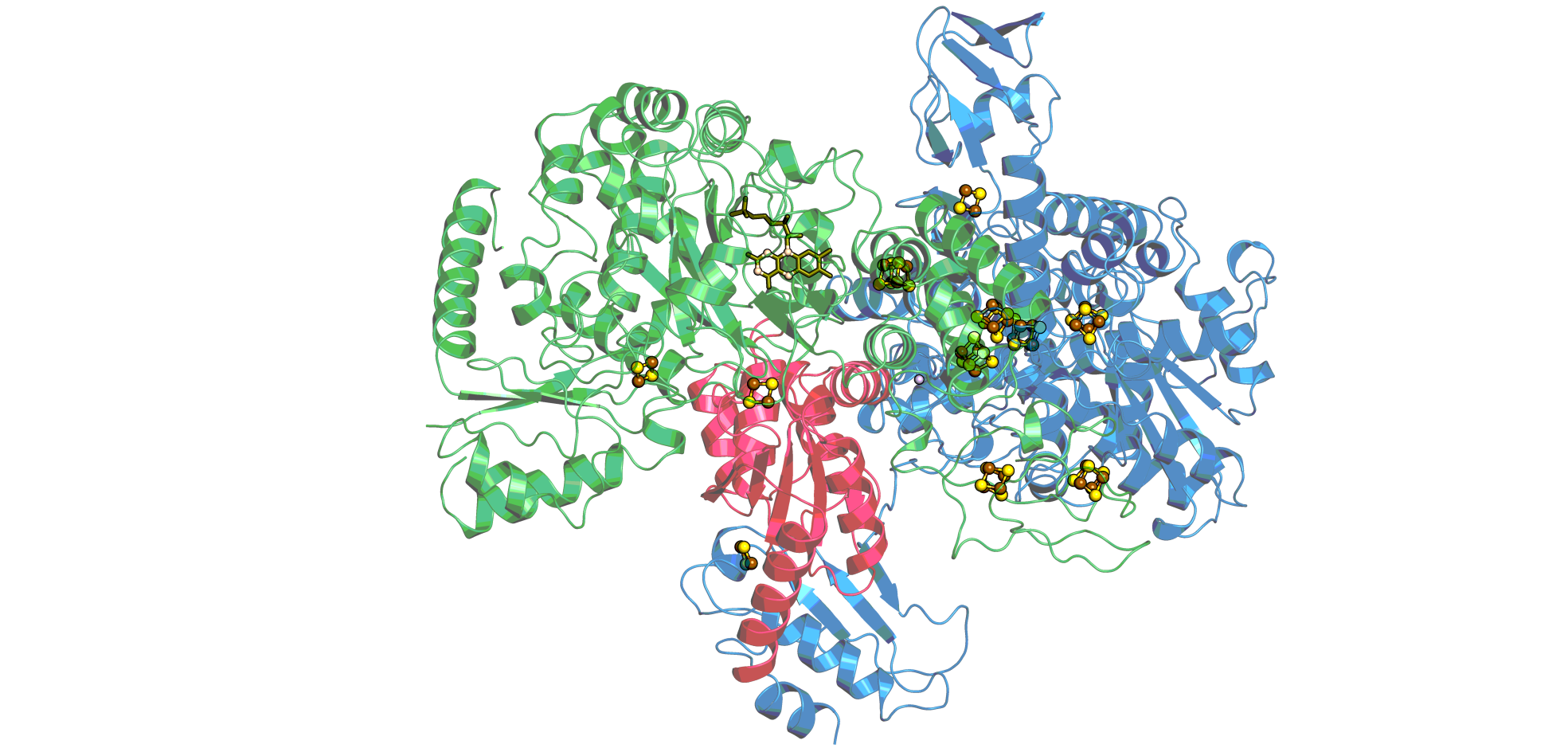
The electron bifurcating hydrogenase (HydABC) from Thermotoga maritimais a multi-centre protein complex containing flavin molecules together with a number of [2Fe-2S]- and [4Fe-4S]-clusters the redox properties of which are not yet well characterized. However, it is the first case of its enzyme family for which a high resolution cryoEM structure has been solved (10.7554/eLife.79361) and which therefore stands out by its strong potential for correlating structural and electrochemical parameters. It has been argued in the past that the ability to bifurcate electrons relies on the specific electrochemical properties of the redox centres forming the bifurcating unit, which, according to the recent structure of the Thermotoga enzyme, involves the flavin as well as one or two [2Fe-2S]- and one [4Fe-4S]-cluster.The collaboration project with the laboratory of James Birrell aims at determining the redox and pH-dependent properties of these centres.
Quinone-based electron bifurctaion occurs at the quinone-oxidizing (Qo) site of Rieske/cytochrome b complexes
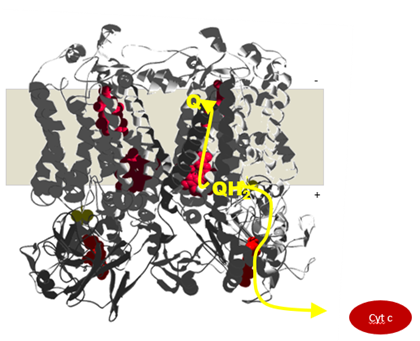
Electrons are transferred to a Rieske iron sulfur cluster (high potential chain) and to a low-potential b-heme (low potential chain). From the low-potential b-heme the electron is transferred across the membrane to another b-heme thereby building-up the proton motive force across the membrane

These reactions rely on a fine-tuned redox potentials of all co-factors:
- the redox midpoint potentials of the low-potential b-heme and the Rieske iron-sulfur cluster are higher and lower than the potential of the two electron transition of the quinone by the same amount.
- The redox potential difference between the two b-heme corresponds to the electrical part of the transmembrane potential
- The redox potential difference between the low potential b-heme and the Rieske iron-sulfur cluster is variable and depends on the species
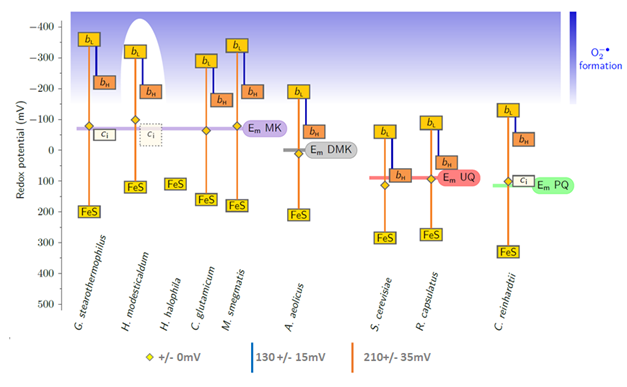
Note that the high redox midpoint potential of ubiquinones and plastoquinones, when compared to menaquinones allowed to take all the cofactros of the complex out of the redox range where reactions with oxygen occur. Only the Qo site semiquinone remains in the dangerous zone.
From low- to high-potential bioenergetic chains: Thermodynamic constraints of Q-cycle function
The modularity of proteins in various bioenergetic chains is of ongoing interest ever since the group was founded. By combining biochemical evidence and phylogenetical analysis we are able to trace back common ancestors and identify patterns that trace back to the protein make-up of the last universal common ancestor (LUCA).
The dyad of the Y-junction- and a flavin module unites diverse redox enzymes
Reactive minerals and the emergence of life
 Our group seeks to understand the evolution of di-iron enzyme reaction centers as found in soluble methane monoxygenase (sMMO) or carbon monoxide dehydrogenase (CODH), and their analgous structural counterparts found in certain reactive naturally occuring minerals such as green rusts, mackinawite, and gregite. Insight into how abiotic geochemical pathways can replicate primative metabolic pathways and self-organize into structures condusive for the continuous production of organic precursors can help us understand how life began it’s inevitable journey to being.
Our group seeks to understand the evolution of di-iron enzyme reaction centers as found in soluble methane monoxygenase (sMMO) or carbon monoxide dehydrogenase (CODH), and their analgous structural counterparts found in certain reactive naturally occuring minerals such as green rusts, mackinawite, and gregite. Insight into how abiotic geochemical pathways can replicate primative metabolic pathways and self-organize into structures condusive for the continuous production of organic precursors can help us understand how life began it’s inevitable journey to being.
Our research:
Methanol on the rocks: Green rust transformation promotes the oxidation of methane
 Coming soon 🙂
Coming soon 🙂
Arsenic Metabolism
The energy substrates used by prokaryotes are extremely diverse and even include substances that are toxic to most organisms. Among them, Arsenic is emerging as a major bioenergetic microbial substrate used since Archean times. Our research objective is not only to understand the place of Arsenic metabolism in the global evolution of energetic processes and life on earth, but also to unravel the still numerous mysteries of the molecular mechanisms of the functioning of the enzymes involved in the bioenergetic metabolism of Arsenic: arsenite oxidase Aio, alternative arsenite oxidase Arx and arsenate reductase Arr. These questions are addressed according to the specificity of our BIP09 team, i.e. the phylogenetic approach combined with biochemistry (enzymology, purifications), biophysics (optical and EPR spectroscopies) and microbiology.
These lead us to study 1) the molecular determinants of the redox and catalytic properties of Molybdenum (Mo) enzymes other than those involved in the metabolism of arsenic, such as TMAO reductase TorA, Soe TtSDH sulphite oxidases, Psr polysuphite reductase, Ttr tetrathionate reductase 2) the phylogeny of enzymes other than those involved in the metabolism of arsenic, in connection with the latter, for example enzymes for the metabolism of sulphur or nitrate, 3) the metabolic interconnexion of arsenic with selenium, sulphur, or photosynthesis.
Arsenic metabolism has even long been proposed as ancient. Arsenic is in fact frequently found in significant concentrations in the vicinity of hot springs, which are considered to mimic the conditions that led to the appearance of life on earth. This hypothesis is now reinforced by experimental paleo-geological data ( 10.1038/ngeo2276). The archaean palaeogeochemical period being anaerobic, the dominant chemical species of arsenic was reduced to arsenite (AsIII). The question is which enzyme may have been responsible for the conversion of this AsIII in the Achean. Two enzymes are known today as being able to oxidize AsIII. The first, arsenite oxidase (Aio; formerly known as Aro, Aso or Aox) is a periplasmic, heterodimeric enzyme with a subunit carrying a Mo-bis-PGD cofactor and a subunit carrying a [2Fe-2S] center called Rieske » (by homology to the Rieske/b complex, see 10.1098/rstb.2002.1184). The phylogeny of the two subunits of Aio, constantly reinforced by new sequences, shows a clear cleavage between Archean and bacterial protein sequences of Aio which suggests that this enzyme was already present in LUCA (10.1093/molbev/msg071, 10.1186/1471-2148-8-206, 10.1016/j.tibs.2008.10.005, 10.1016/j.bbabio.2012.08.007), thus reinforcing the hypothesis of the ancestrality of this enzyme.
Despite the resolution of Aio’s structures, the question of how the enzyme works has not yet been resolved. Our hypothesis is that the two electrons from the MoIV formed during the oxidation of AsIII are transferred simultaneously to the two [FeS] centers during an electron bifurcation mechanism. This hypothesis is compatible with the internal electron transfer kinetics measured on the wild type enzyme and the redox properties of the enzyme (10.1016/j.bbabio.2017.08.003, 10.1016/j.bbabio.2016.05.003). We are, however, currently testing this hypothesis by characterizing mutants of the enzyme.
Characterization of several representatives of the enzyme revealed that the Aio from proteobacteria (10.1074/jbc.M110.113761 )constitute two distinct enzyme groups in terms of redox partner specificity. One reacts with horse heart cytochrome, while the other is unable of doing so, but reacts for example with Aquifex aeolicus cytochrome c555 or Azurin. Analyses of surface electrostatic potentials of enzymes and electronic carriers suggest that it is the electrostatic compatibility between enzyme and electronic carrier that is at the origin of the specificities observed. For example, Azurin and the cytochrome c555 of Aquifex aeolicus react with the Aio from A. faecalis and not that from NT-26. The reverse is also true for horse heart cytochrome.
The second enzyme known to oxidize AsIII is the alternative arsenite oxidase Arx. This enzyme is a very close homologue of the arsenate (AsV) reductase Arr. Arr is a heterotrimeric enzyme also with Mo-bis-PGD cofactor, which functions as a terminal reductase using AsV as an electron acceptor, drawing its electrons from a pool of membrane quinones through the operation of its membrane subunit. By combining phylogenetics of arr/arx genes, enzymology and microbiology, our work suggests that Arx and Arr have recently evolved in the living world and that Arx has been very poorly distributed (10.1186/1471-2148-8-206, 10.1126/science.1164967, 10.1016/j.bbabio.2012.08.007; 10.1016/j.bbabio.2020.148252). Our interpretation of this phylogeny is that only after the diversification of Archaea and Bacteria, O2 levels induced an accumulation of the oxidized form of Arsenic, AsV, leading to the appearance of Arr in bacteria. Thermodynamic considerations, confirmed by experimental data, suggest that Arr reduces AsV by oxidizing menaquinones (MK, quinones of relatively low redox potential), as in the Shewanella ANA-3 bacterium, while Arx reduces ubiquinone by oxidizing AsIII (10.1016/j.bbabio.2020.148252). This hypothesis is consistent with the distribution of quinones in the tree of life. According to the phylogeny of the enzymes at Mo, the enzymes Arx and Arr seem to have evolved from an ancestral enzyme involved in sulphur metabolism (10.1016/j.bbabio.2020.148279).
The family of Mo enzymes is one of the richest enzyme families in biology (10.1039/9781782623915-00143, 10.1016/j.bbabio.2013.01.011) and probably one of the oldest in the living world (10.1038/srep00263). There are several families of enzymes carrying Molybdenum cofactor (pterin-free, with one pterin, with two pterins). The team is mainly studying two-pterins enzymes known as Mo-bisPGD enzymes, exclusively found in prokaryotes.
The first question addressed on this family of enzymes is the diversity of their catalytic capacity, which is without any equivalence in the enzymatic world. Despite 30 years of research, the question of the structural determinants behind the catalytic properties of these enzymes has not been resolved. The question is addressed in the team, using the phylogenetic approach, in two groups of Mo-enzymes. The aim is to determine the amino acids responsible for the enzymatic specificities by bioinformatic analysis (structure and multiple sequence alignment) of the enzymes (description of the approach in 10.1039/9781782623915-00143). The sub-family we are currently working on is the Psr/Phs-Ttr-Arr/Arx group, comprising enzymes capable of converting arsenic and others capable of converting sulphur.
The adaptation of Mo enzymes to their substrate is also correlated with the adaptation of the redox properties of its Mo-bis-PGD cofactor. Here again, the questions of the structural origin of the redox plasticity of this center remain unanswered. Analyzing the structure of Aio and other Mo-enzymes, stabilizing a MoV or not state, suggested the crucial role of the hydrogen bond network around Mo and particularly with nitrogen 5 of one of the two pterins at the origin of the redox properties of the cofactor (10.1016/j.bbabio.2016.05.003). We are currently testing this hypothesis with the Thermus thermophilus sulphite dehydrogenase.
Rieske/cytochrome b complexes
Rieske/cytb complexes get their names from the two conserved protein subunits. A transmembrane subunit with two b-type hemes and an iron-sulfur subunit, named after the M Rieske who first described it. These two submits and the quinol-binding site they form are responsible for the catalytic reaction of the enzyme shuttling electrons from quinol to cytochrome c. Each electron crosses the membrane before continuing to cytochrome c and thus contributes to the build-up of the proton motive force.
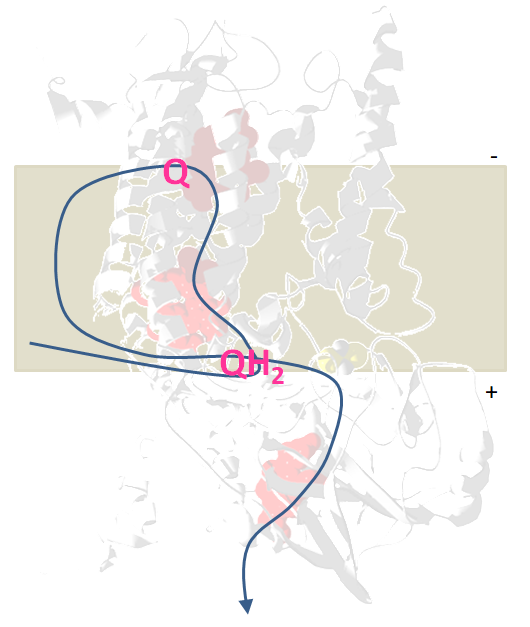
Electron bifurcation in the quinol oxidizing site confers to one of the electrons from the quinol a high redox potential, it will reduce the iron-sulfur cluster and cytochrome c. The second electron gains a much more negative redox potential enabling it to cross the membrane against the gradient and reduces a quinone.
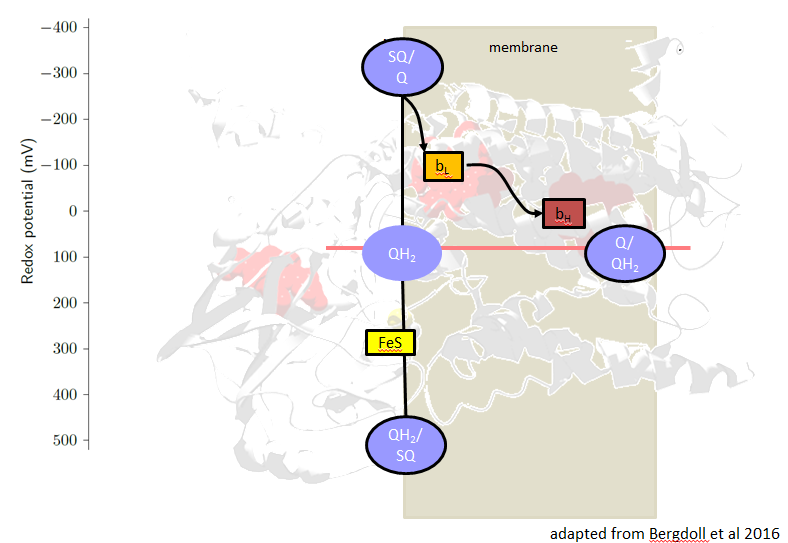
The redox midpoint potentials of all cofactors adapt to the quinone potential, the potentials of heme bL and the [2Fe-2S]-cluster are arranged symmetrically around the quinone potential. The potential difference between the hemes bL and bH corresponds to the DY of the pmf.
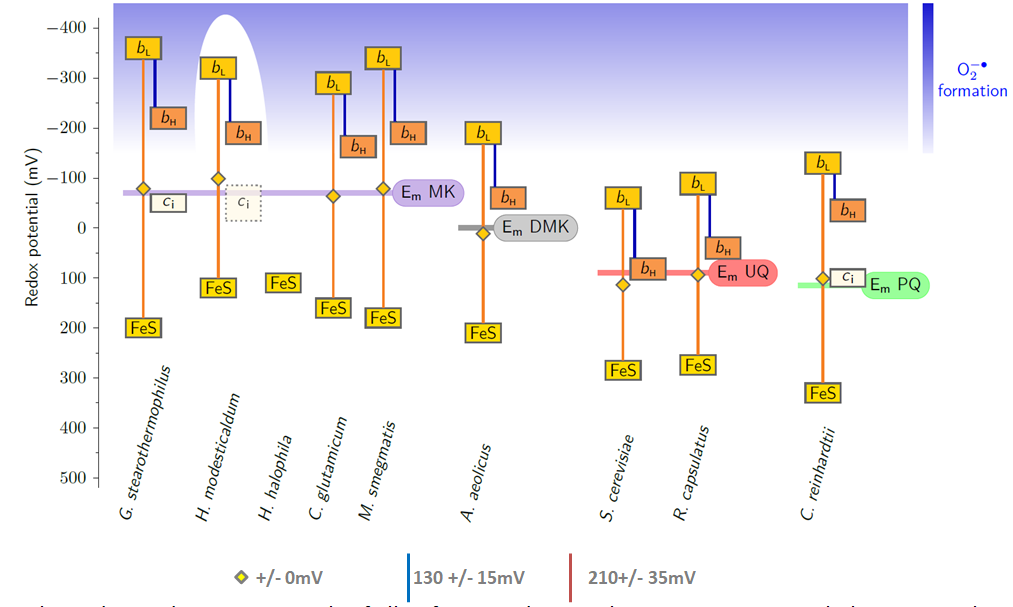
Rieske/cytb complexes can be found in bioenergetic reaction chains with enough redox energy to transfer several electrons over the membrane.
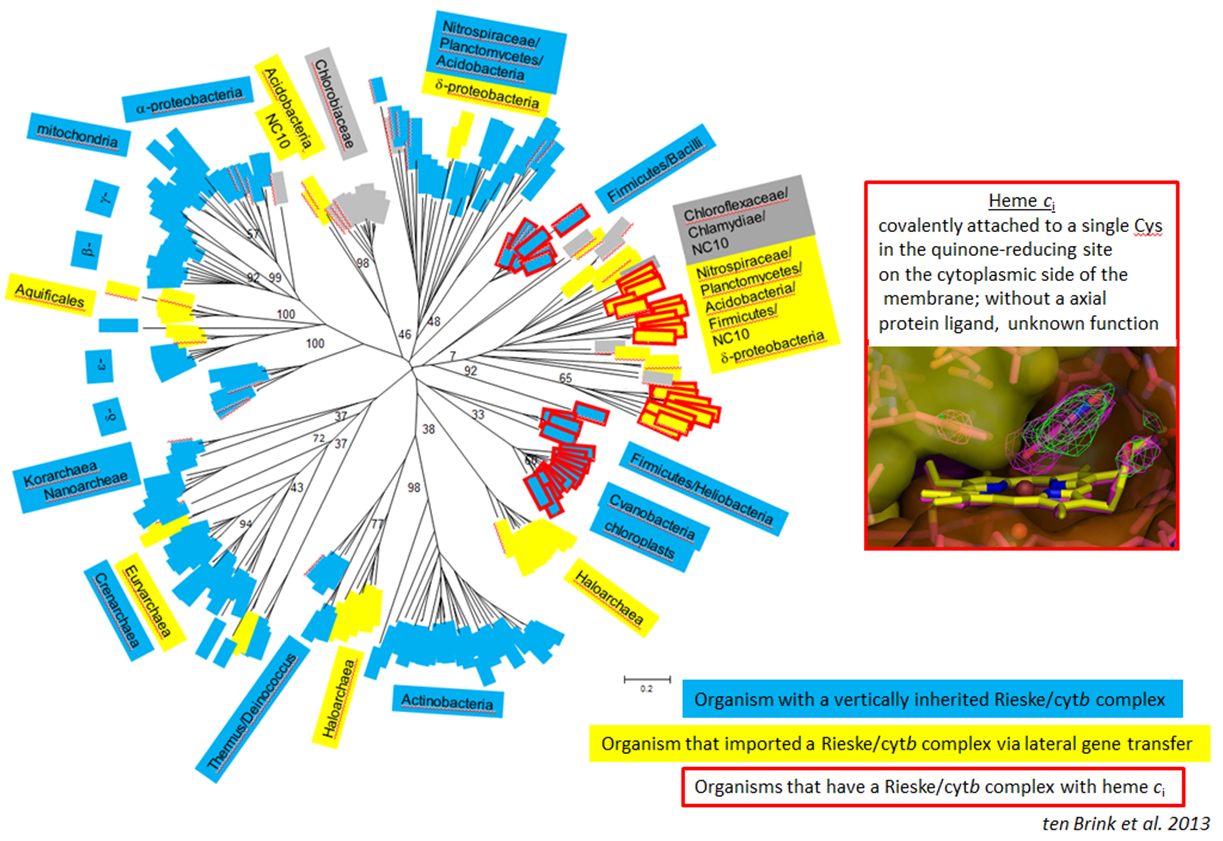
Rieske/cytb complexes can be found in most Phyla and the phylogenetic tree is compatible with a mainly vertical inheritance since the Last Universal Common Ancestor. In come enzymes an additional heme cofactor can be found. This heme is unusual, since it is a c-type heme, attached to the protein with a single cysteine and located in a quinone-binding site on the negative side of the membrane. Complexes with this co-factor are present in Firmicutes, cyanobacteria and chloroplasts and the enzyme has been laterally transferred to a wide variety of species among which are anammox bacteria.